Algorithms
Microscopy Image Restoration and Segmentation
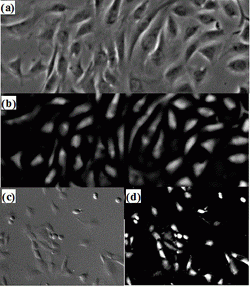
Based on the principle of detection-based tracking approach (i.e. first detect, then track), segmenting object candidates from the background is fundamental to the tracking system. Figure 1(a) shows a typical phase contrast image where cells are surrounded by bright halos, some cell structures (e.g. nuclei and cell membrane) consist of multiple dark regions, and cellular fluid inside the membrane has similar intensity as the background. This presents a lot of challenges to traditional image segmentation techniques. Instead, we attack the segmentation problem by considering the unique characteristics of microscopy image formation process such as the halo artifacts of phase contrast images. Using a recently discovered microscopy imaging model, we formulate a regularized quadratic cost function to restore the artifact-free microscopy images: Figure 1(b) shows the restored image where a simple thresholding method is enough to separate the cell pixels from background pixels. Differential Interference Contrast (DIC), another popular microscopy imaging technique, displays a pseudo three dimensional shading effect and cell intensities are quite similar to background pixels as shown in Figure1(c). Similarly, based on the DIC imaging model, we can restore an image conducive to thresholding (Figure1(d)).
Mitosis Detection
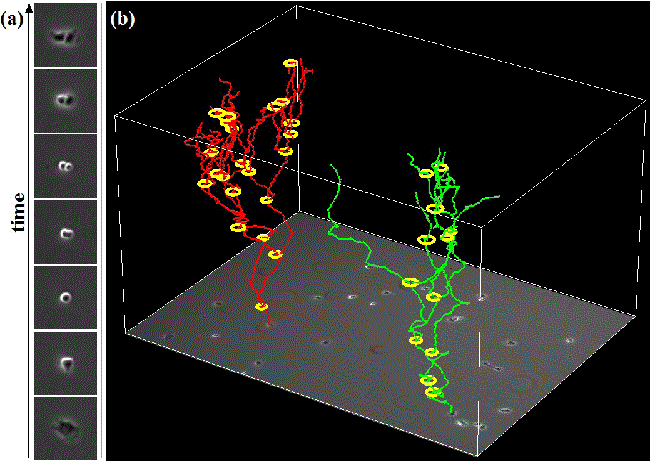
We propose an effective approach for automated mitosis detection using phase-contrast time-lapse microscopy, which is a non-destructive imaging modality, thereby allowing continuous monitoring of cells in culture. In our approach, we present a probabilistic model for event detection, which can simultaneously 1) identify spatio-temporal patch sequences that contain a mitotic event and 2) localize a birth event, defined as the time and location at which cell division is completed and two daughter cells are born. Our approach significantly outperforms previous approaches in terms of both detection accuracy and computational efficiency, when applied to multipotent C3H10T1/2 mesenchymal and C2C12 myoblastic stem cell populations.
Cell Tracking based on Frame by Frame Data Association
The image segmentation algorithm segments blobs from input images that can be individual cells or cell clusters (overlapped cells), and the mitosis detection algorithm locates
birth events where and when one cell divides into two cells. Based on the outputs of these two algorithms, we developed a cell-blob correspondence algorithm performing
data association between the cells in the previous frame and the blobs segmented in the current frame. In detail, the association algorithm makes the following hypotheses with
corresponding likelihood for all possible cell actions:
- one-to-one hypothesis: a cell migrates to a new position.
- one-to-none hypothesis: a cell exits from the field of view. Only cells near the image boundary have one-to-none (exit) hypotheses.
- none-to-one hypothesis: a blob enters the field of view:Similar to one-to-none hypothesis, only blobs near the image boundary have none-to-one (enter) hypothesis.
- one-to-two: a cell divides into two cells. Only cells near the spatiotemporal location of a birth event have one-to-two (mitosis) hypothesis.
- many-to-one: multiple cells overlap. When several cells are close to each other in the previous frame and a nearby large blob is detected in the current frame, those cells have a many-to-one hypothesis. When tracking the overlapped blob in the succeeding frames, we apply a contour-matching method to separate it into its member cells, thus the cell identities are maintained.
Cell Tracking based on Global Data Association. [see paper]
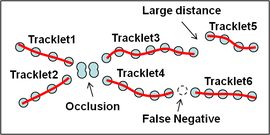
Automated cell tracking in populations is important for research and discovery in biology and medicine. We propose a cell tracking method based on global spatiotemporal data association which considers hypotheses of initialization, termination, translation, division and false positive in an integrated formulation. Firstly, reliable tracklets (i.e., short trajectories) are generated by linking detection responses based on frame-by-frame association. Next, these tracklets are globally associated over time to obtain final cell trajectories and lineage trees. During global association, tracklets form tree structures where a mother cell divides into two daughter cells. We formulate the global association for tree structures as a maximum-a-posteriori (MAP) problem and solve it by linear programming. This approach is quantitatively evaluated on sequences with thousands of cells captured over several days.
Videos